What I do have some idea about however is the end result of X-ray crystallography; a model of a protein molecule or a nucleic acid– a very, very detailed model.
In the very best models, consisting of hundreds or thousands of amino acids, single atoms of each amino acid can be discerned – giving an amazingly complex model of the protein totaling tens of thousands of atoms. Navigating and orientating yourself around such a vast and detailled model is difficult enough, but producing such a thing is far, far more difficult. So, why would you want to expend so such time and effort on producing such a model of a protein or a nucleic acid?
Producing these models - although innately intricate and beautiful - isn't an end in itself. These models are capable of telling us vast amounts of information on how the proteins work and interact with other proteins. A detailed-enough model can describe the mechanism of an enzyme's catalysis to the atom; something which reminds me of how astounding this field of science is. There is something both unsettling and supremely comforting when a process is understood in such detail that we understand it on the most fundamental level.
 |
| Ribosome structure determined by X-ray crystallography from Venki Ramakrishnan |
In the very best models, consisting of hundreds or thousands of amino acids, single atoms of each amino acid can be discerned – giving an amazingly complex model of the protein totaling tens of thousands of atoms. Navigating and orientating yourself around such a vast and detailled model is difficult enough, but producing such a thing is far, far more difficult. So, why would you want to expend so such time and effort on producing such a model of a protein or a nucleic acid?
Producing these models - although innately intricate and beautiful - isn't an end in itself. These models are capable of telling us vast amounts of information on how the proteins work and interact with other proteins. A detailed-enough model can describe the mechanism of an enzyme's catalysis to the atom; something which reminds me of how astounding this field of science is. There is something both unsettling and supremely comforting when a process is understood in such detail that we understand it on the most fundamental level.
However, achieving these models is anything but easy. A whistle-stop tour towards a model would go something like this;
Firstly, there is the matter of producing enough protein in order to examine it via crystallography. This involves (normally) knowing the genetic sequence, and implanting the gene into our good old friend E.coli. There's all sorts of issues here, such as your protein falling apart (degradation), or clumping together (precipitation/aggregation) or the protein simply being insoluble.
Secondly, there's the small matter of purifying the protein. There's a few thousand proteins in E.coli which you won't want to look at, and so finding a method of extracting your protein of interest is an art in itself. Methods generally rely on the concept of chromatography, a method of separating a mixture of stuff based on its affinity to something else. Being able to produce a large amount of pure protein isn't simple nor easy, and this step alone can take an inordinately large amount of time.
 |
| Pretty... |
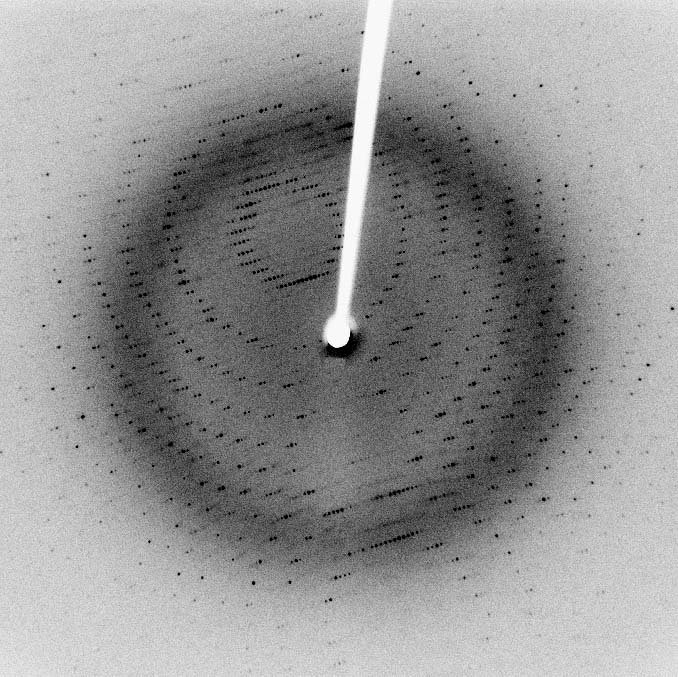 |
| Diffraction patterns aren't particularly accessible. |
It's not a particularly romantic process, and it's not really completely comprehensible. But somewhere between the complex maths, and the thousands of crystallisation trials, the end product seems justified.
No comments:
Post a Comment